Our research focuses on diffusion magnetic resonance imaging, the only method that can map the connections of the living human brain. Our ongoing research includes NIH R01 projects to enable the analysis of extremely large datasets and to map previously invisible superficial white matter. We are also involved in projects related to analysis of diffusion tractography via fiber clustering, creation of white matter atlases, extraction of machine-learning-relevant features from the white matter, and development of open-source software. We have a longstanding collaboration with Dr. Alexandra J. Golby on topics related to neurosurgical planning.
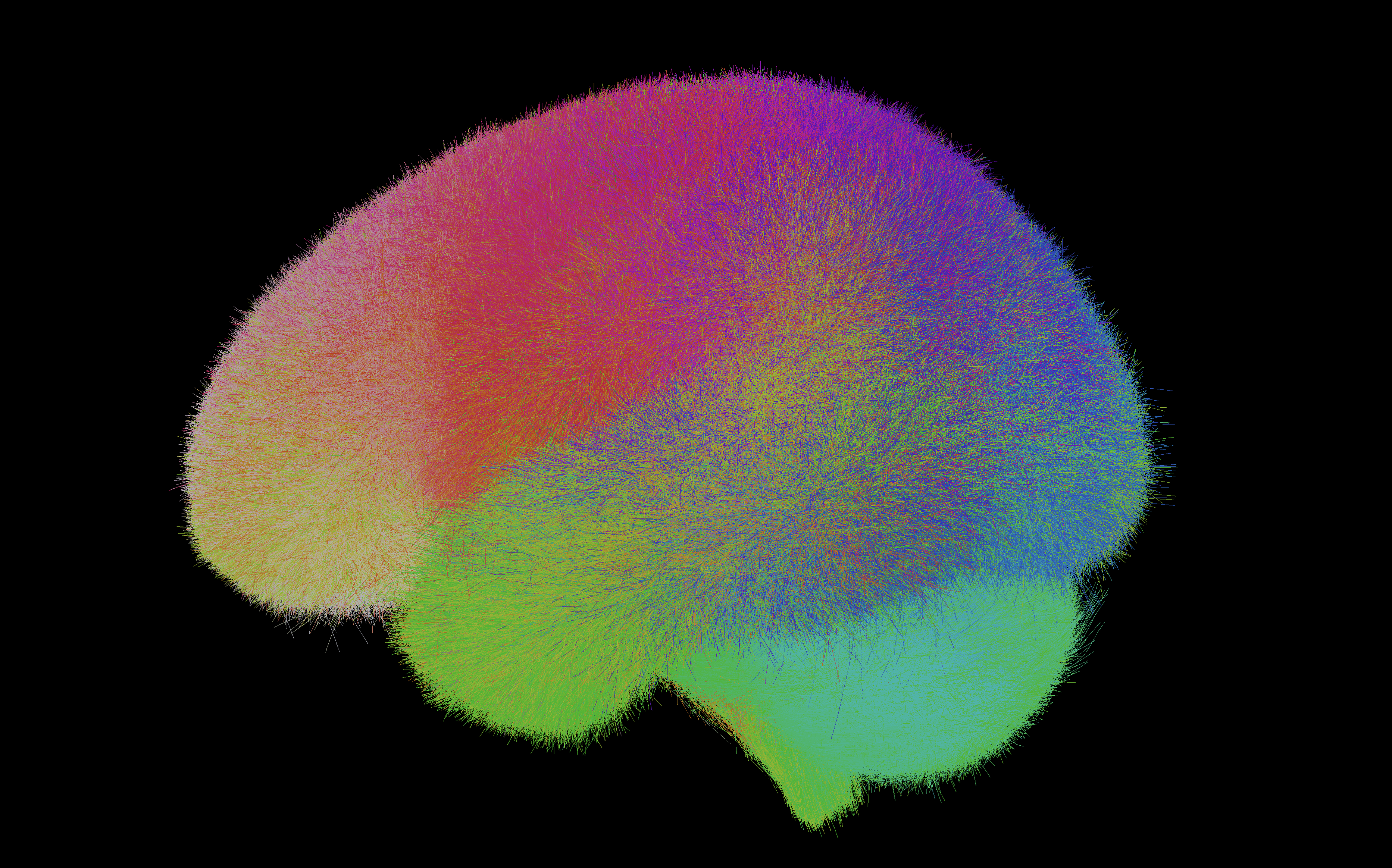
White Matter Atlases
One research direction of the O’Donnell Laboratory is the creation of white matter atlases using a combination of machine learning methods and expert neuroanatomical curation. We created an anatomically curated white matter atlas to enable consistent white matter tract parcellation across different populations (Zhang et al. 2018). The atlas contains 58 deep white matter tracts including major long-range association and projection tracts, commissural tracts, and tracts related to the brainstem and cerebellar connections, plus 198 short and medium-range superficial fiber clusters organized into 16 categories according to the brain lobes they connect. See more information and atlas download instructions.
Mapping Superficial White Matter
NIH R01 MH125860
This 5-year R01 project (PIs: Lauren J. O’Donnell, Yogesh Rathi, Nikos Makris) is titled “Mapping the Superficial White Matter Connectome of the Human Brain Using Ultra High Resolution Multi-Contrast Diffusion MRI”. We propose to create the first atlas of the human brain’s superficial white matter (SWM) using sub-millimeter ultra high resolution diffusion MRI (dMRI). The SWM is located between the deep white matter and the cortex. Our understanding of human neuroanatomy relies heavily on the results of invasive tracer studies in monkeys, but the detailed neuroanatomy of the SWM in monkeys has not yet been systematically compiled or analyzed. We propose to address these challenges to create the most comprehensive description of the SWM to date.
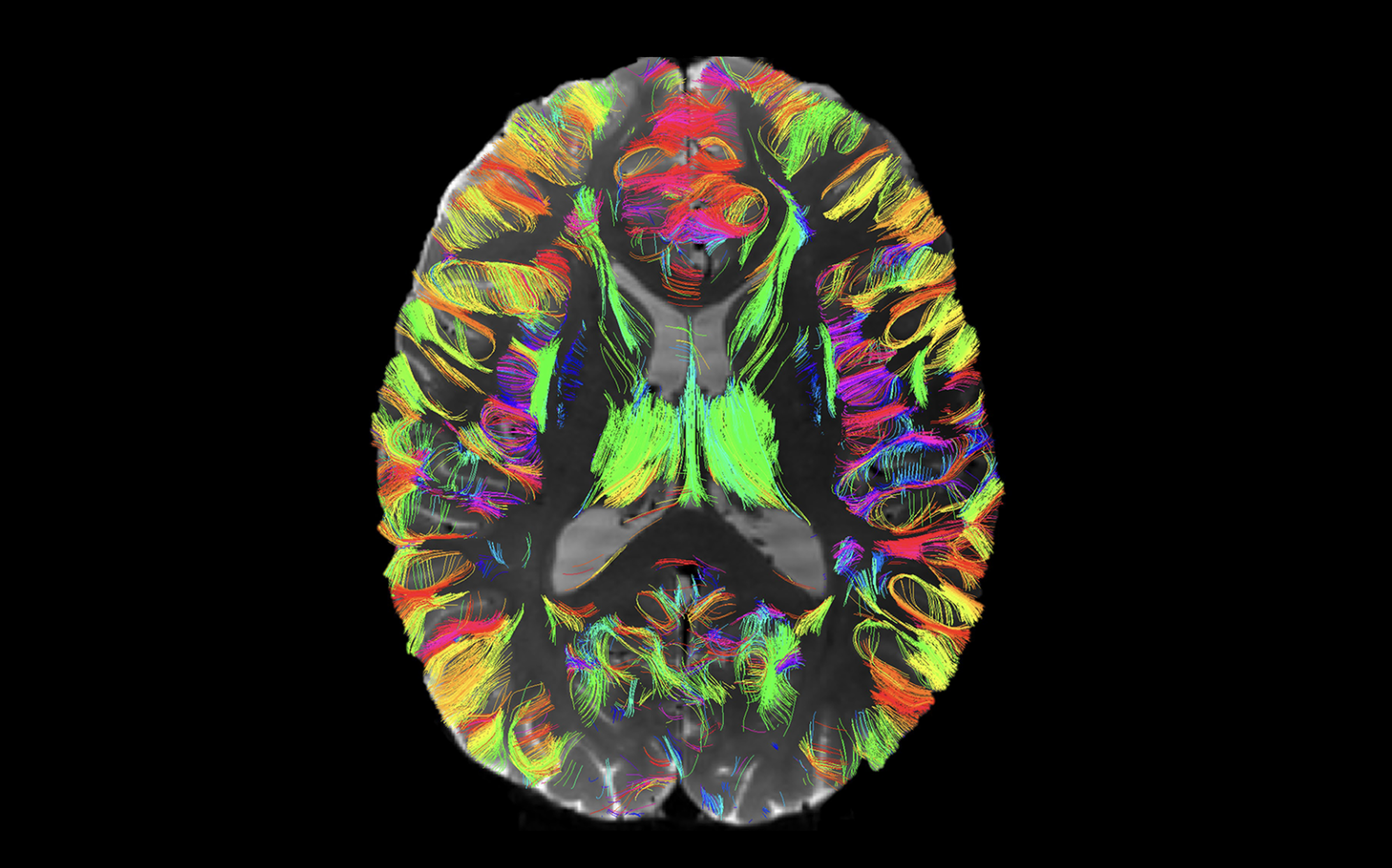

Mapping Cerebellar White Matter
NIH R01 MH132610
This 5-year R01 project (PIs: Lauren J. O’Donnell, Yogesh Rathi, Nikos Makris) titled “Mapping of the Intrinsic and Extrinsic Cerebellar Connectome at Ultra High Resolution With Expert Neuroanatomical Curation”. We propose to create the first detailed atlas of the human cerebellar connectome using sub-millimeter ultra-high-resolution diffusion MRI (dMRI). The complex anatomy of the cerebellar structural connectome includes intricate connections between the tightly foliated cerebellar cortex, the deep cerebellar nuclei, and structures external to the cerebellum including the spinal cord, brainstem, thalamus, and cerebral cortex. Our final deliverable will be a comprehensive, anatomically curated atlas of the human cerebellar connectome, which will enable the study of the cerebellar connectome in health and disease.
SlicerDMRI Open-Source Software
Our NIH-funded open-source software, SlicerDMRI, is used in multiple brain research studies for diffusion MRI visualization and analysis. Our software for diffusion MRI fiber tractography clustering, whitematteranalysis, is available as open source. In addition, our laboratory has developed a variety of open-source software, including state-of-the-art deep learning methods.

Diffusion MRI Data Harmonization
NIH R01 MH119222
This 5-year R01 project (PIs: Yogesh Rathi and Lauren J. O’Donnell) is titled “Harmonizing Multi-Site Diffusion MRI Acquisitions for Neuroscientific Analysis Across Ages and Brain Disorders”. Several large studies such as the Human Connectome Project (HCP) and the Adolescent Brain Cognitive Development (ABCD) have collected or are poised to collect diffusion MRI data from over 30,000 subjects. However, an important challenge is that these datasets collected from different scanners cannot be pooled for joint analysis due to large inter-scanner (inter-site) differences. In this grant, we propose to address these challenges to enable large-scale data-intensive analysis of dMRI data. We recently created a dataset of harmonized and processed ABCD dMRI data (from release 3), comprising quality-controlled imaging data from 9,345 subjects, focusing exclusively on the baseline session, i.e., the first time point of the study. This resource required substantial computational time (approx. 50,000 CPU hours) for harmonization, whole-brain tractography, and white matter parcellation. The dataset includes harmonized dMRI data, 800 white matter clusters, 73 anatomically labeled white matter tracts in full and low resolution, and 804 different dMRI-derived measures per subject (72.3 TB total size). Accessible via the NIMH Data Archive, it offers a large-scale dMRI dataset for studying structural connectivity in child and adolescent neurodevelopment.
Cetin-Karayumak S, Zhang F, Zurrin R, Billah T, Zekelman L, Makris N, Pieper S, O’Donnell LJ, Rathi Y. Harmonized diffusion MRI data and white matter measures from the Adolescent Brain Cognitive Development Study. Scientific Data. 2024 Feb 27;11(1):249.